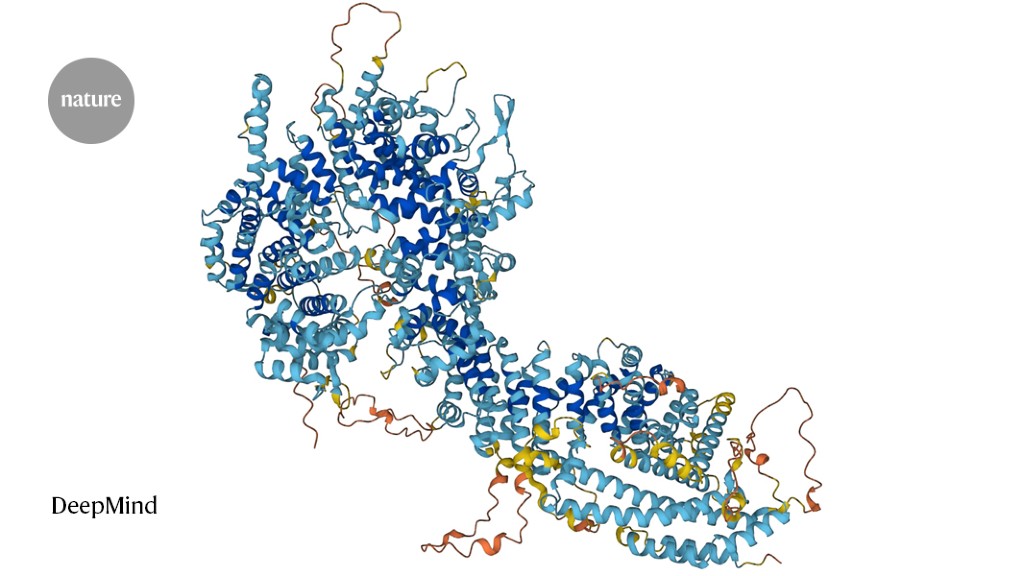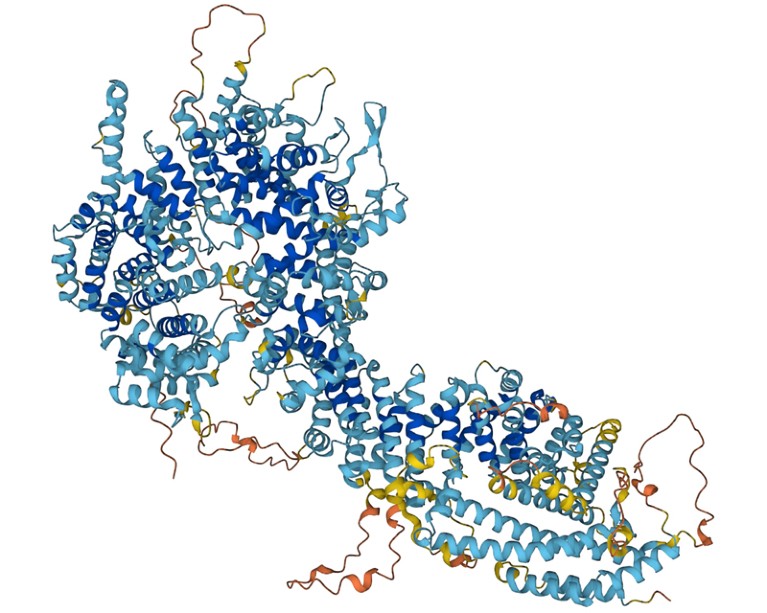
Foldseek permits researchers to determine proteins whose form resembles that of different proteins.Credit score: DeepMind
Once you uncover a protein, how do you identify what it does? That’s the issue Gregory Gloor was going through.
A biochemist on the College of Western Ontario in London, Canada, Gloor was learning bacterial communities at an oil-refinery wastewater remedy plant, hoping to determine the proteins that assist them to degrade poisonous substances. As a proof of idea, he began trying on the proteins expressed by viruses referred to as bacteriophages that infect these micro organism. Sadly, a search of databases of recognized proteins for matches got here up empty.
Then Gloor realized of a search instrument referred to as Foldseek, first shared by its creators in 2021 and described in Could in Nature Biotechnology1. “It was like, hallelujah,” he says. His mission “went from mainly unimaginable to doable”.
Proteins are constructed of chains of amino acids, and their folded form dictates their perform. Previously few years, artificial-intelligence instruments that predict a protein’s 3D construction from its amino-acid sequence alone — versus figuring out that construction experimentally — have improved drastically. Researchers have used AlphaFold 2, from Google DeepMind in London; RoseTTAFold, from a group on the College of Washington, Seattle; and different such instruments to compile databases containing a whole bunch of hundreds of thousands of constructions. Foldseek makes it doable to rapidly search these databases for proteins which have related shapes — and presumably, related capabilities — to a protein of curiosity.
Better of each worlds
The standard computational method to figuring out the perform of an unfamiliar protein is to search for proteins with related amino-acid sequences. If the capabilities of these associated proteins are recognized, researchers could make a guess as to what the brand new protein would possibly do.
Sequence searches are quick, like looking a tough drive for a file identify. However they typically miss good matches as a result of proteins with related shapes can have vastly completely different sequences. Construction-based search strategies search for shapes as an alternative of sequences, however this could take hundreds of occasions longer, as a result of it’s computationally troublesome to match complicated 3D objects. With Foldseek, researchers obtained one of the best of each worlds: the software program represents a protein’s form as a string of letters — a ‘structural alphabet’ — thereby providing the sensitivity of shape-based searches however on the velocity of sequence-based ones.
“One of many key concepts was that in an effort to produce an excellent structural search, you will need to get the encoding proper,” says Martin Steinegger, a biologist at Seoul Nationwide College and one of many Foldseek paper’s lead authors.
What’s subsequent for AlphaFold and the AI protein-folding revolution
Gloor used ColabFold, a cloud-based computational-notebook interface to AlphaFold 2, to foretell the constructions of the bacteriophage proteins he discovered, after which Foldseek to match them to recognized proteins. A few of the proteins, he discovered, fashioned the viruses’ outer shells; others had been enzymes2. His evaluation: Foldseek is “amazingly intelligent”.
Foldseek is just not the primary algorithm to cut back protein construction to an alphabet. Different search instruments sometimes assign every amino acid a letter on the premise of its orientation relative to the amino acids instantly earlier than and after it within the protein sequence. Nonetheless, that method overlooks interactions between amino acids which are far aside within the linear chain, however close by in 3D house. Foldseek assigns every amino acid one in every of 20 letters, on the premise of its distance from, and orientation relative to, the amino acid that’s closest within the folded-up protein. By specializing in these spatial bridges, Steinegger says, Foldseek’s ‘3D-interaction alphabet’ higher captures world construction.
Seeing again in time
“Biology happens in three dimensions,” says Janet Thornton, a computational biologist on the European Molecular Biology Laboratory’s European Bioinformatics Institute in Hinxton, UK. The power to match proteins on the premise of their form “lets you see a lot farther again in evolutionary time, which lets you determine very distant family members that developed from the identical precursor” protein, she says.
To check Foldseek, Steinegger’s group used a database of 365,000 proteins whose shapes had been predicted utilizing AlphaFold 2. They fed 100 of those shapes into Foldseek and requested it to rank, for every one, essentially the most related proteins within the database. The rating was based mostly on what number of ‘true positives’ the algorithm retrieved (that’s, proteins scoring above a sure similarity threshold in response to atomic modelling) earlier than retrieving a false constructive. Foldseek outperformed two well-liked structure-based search instruments, TM-align and Dali — performing 24% and eight% higher, respectively — and practically 35,000 and 20,000 sooner. In contrast with a structural-alphabet-based instrument referred to as CLE-SW, Foldseek was 23% higher, and 11 occasions as quick1.
DeepMind’s AI predicts constructions for an unlimited trove of proteins
Foldseek is obtainable as open-source software program for macOS and Linux computer systems. The builders additionally created an internet server for researchers to go looking any of seven structural databases masking a whole bunch of hundreds of thousands of proteins. In keeping with Steinegger, the software program has been put in no less than 14,000 occasions, and researchers run about 300 searches on the server every day.
Thornton says Foldseek may assist researchers to determine protein capabilities in new pathogens, or just make clear how organisms function. For instance, Steinegger and his group utilized Foldseek to search out clusters of associated proteins within the AlphaFold database and recognized bacterial proteins with an analogous construction to a human histone3.
As for Gloor, with current search instruments, he discovered matches for less than a small fraction of the bacteriophage proteins in his examine, none of which had recognized capabilities. Utilizing Foldseek, he discovered matches for half of his proteins, figuring out 15% as enzymes2.
“Changing a three-dimensional quantity of interactions down right into a string required a good bit of perception and originality,” Gloor says. And utilizing Foldseek, scientists can perceive many extra proteins in lots of extra organisms. “It’s actually going to vary the way in which that we do evolutionary research,” he says. “It can improve our capability to look in actually distinctive ecosystems and determine how they work.”